Assumptions
Last Updated: 03, December, 2024 at 09:21
- Read some data
- Assumption 1: Linearity of model
- Assumption 2: Normal distribution of the errors
- Assumption 3: Homoscedasticity
- More examples
Read some data
library(tidyverse)
## ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.2 ──
## ✔ ggplot2 3.4.0 ✔ purrr 1.0.2
## ✔ tibble 3.1.8 ✔ dplyr 1.0.10
## ✔ tidyr 1.2.1 ✔ stringr 1.4.1
## ✔ readr 2.1.3 ✔ forcats 0.5.2
## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
body_data <-read_csv('data/body.csv')
## Rows: 507 Columns: 25
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## dbl (25): Biacromial, Biiliac, Bitrochanteric, ChestDepth, ChestDia, ElbowDi...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
vik_data <-read_csv('data/vik_table_9_2.csv')
## Rows: 12 Columns: 4
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## dbl (4): Person, Y, X1, X2
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Assumption 1: Linearity of model
Example 1
plot(vik_data$X1, vik_data$Y)
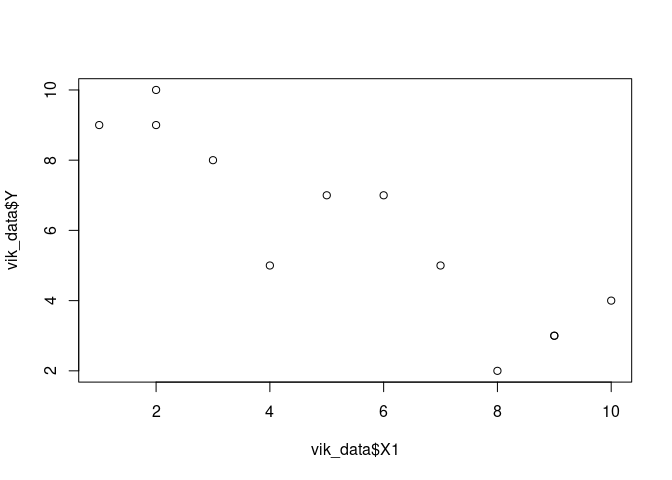
model <- lm(Y ~ X1, data = vik_data)
plot(fitted(model), resid(model))
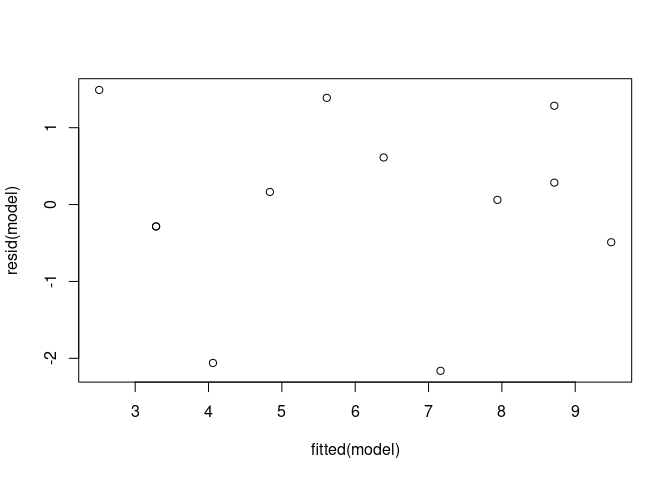 Let’s
create the diagnostic plots. For our purposes, we will only look at the
top two plots.
Let’s
create the diagnostic plots. For our purposes, we will only look at the
top two plots.
# Split the plotting panel into a 2 x 2 grid. Such that we get the four plots
# in 4 separate panels.
par(mfrow = c(2, 2))
plot(model)
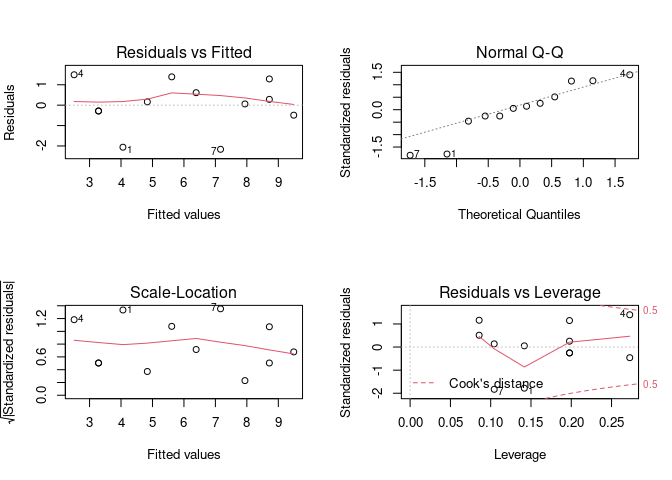
# We can reset the plotting panel using this code: par(mfrow = c(1, 1))
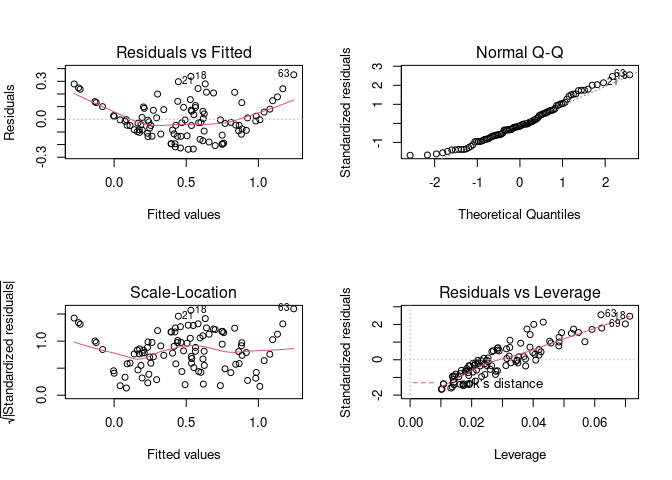
Example 2
x_data <- runif(100)
y_data <- x_data^3 + rnorm(100, sd=0.05)
fake <- tibble(x_data=x_data, y_data = y_data)
model <- lm(y_data ~ x_data, data = fake)
par(mfrow = c(1, 1))
plot(x_data, y_data)
abline(model, col='red')
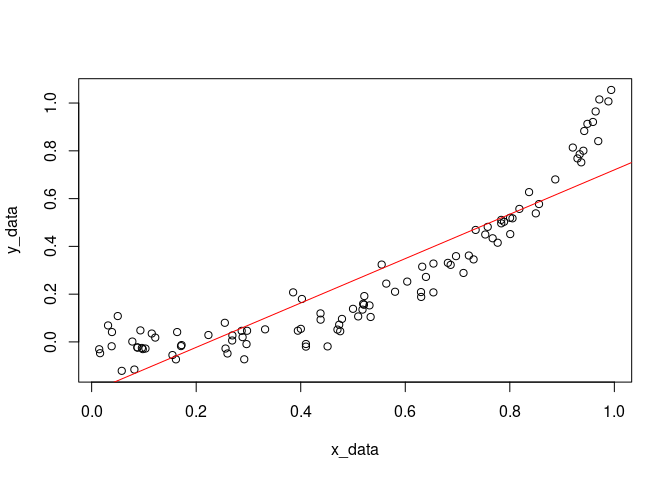
model <- lm(y_data ~ x_data, data = fake)
summary(model)
##
## Call:
## lm(formula = y_data ~ x_data, data = fake)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.22963 -0.10576 -0.01829 0.09487 0.34005
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -0.20838 0.02650 -7.862 4.99e-12 ***
## x_data 0.92877 0.04503 20.626 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.1346 on 98 degrees of freedom
## Multiple R-squared: 0.8128, Adjusted R-squared: 0.8109
## F-statistic: 425.4 on 1 and 98 DF, p-value: < 2.2e-16
par(mfrow = c(2, 2)) # Split the plotting panel into a 2 x 2 grid
plot(model)
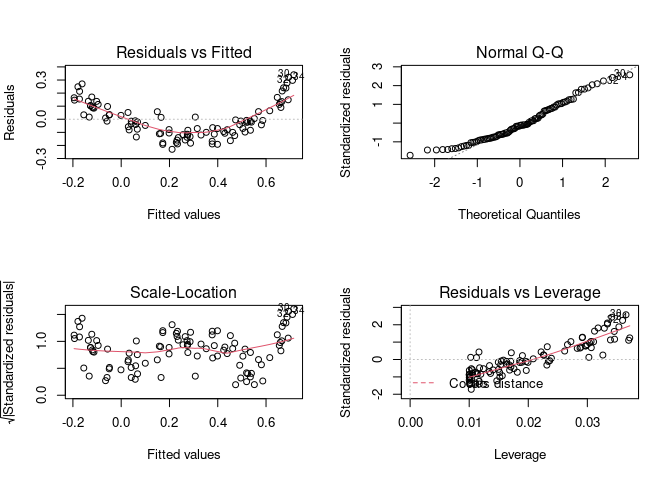
par(mfrow = c(1, 1))
Let’s run the model on transformed data and look at the diagnostic plots again.
model <- lm(y_data ~ I(x_data^3), data = fake)
par(mfrow = c(2, 2)) # Split the plotting panel into a 2 x 2 grid
plot(model)
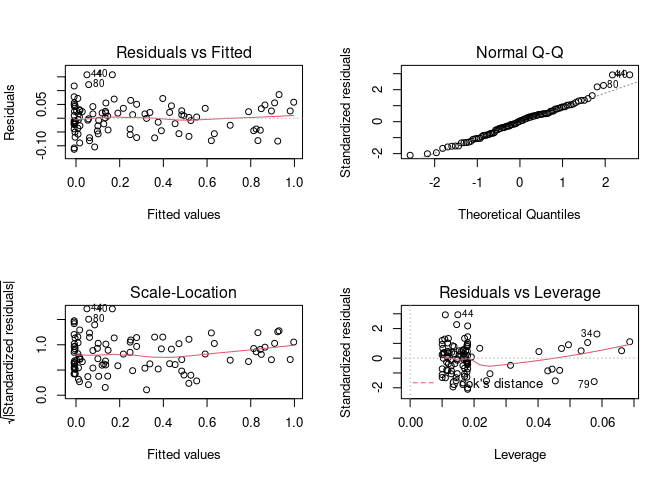
par(mfrow = c(1, 1))
Example 3
This demonstrates that the diagnostic plots can also be used when we have multiple predictors (in this case plotting the dependent vs the independents might be difficult or impossible).
x1_data <- runif(100)
x2_data <- runif(100)
y_data <- x1_data^3 + x2_data^3
fake <- tibble(x1_data=x1_data,x2_data=x2_data, y_data = y_data)
model <- lm(y_data ~ x1_data + x2_data, data = fake)
par(mfrow = c(2, 2)) # Split the plotting panel into a 2 x 2 grid
plot(model)
par(mfrow = c(1, 1))
Example 4
Finally, an example with real data.
model <- lm(Weight ~ Height, data = body_data)
par(mfrow = c(2, 2)) # Split the plotting panel into a 2 x 2 grid
plot(model)
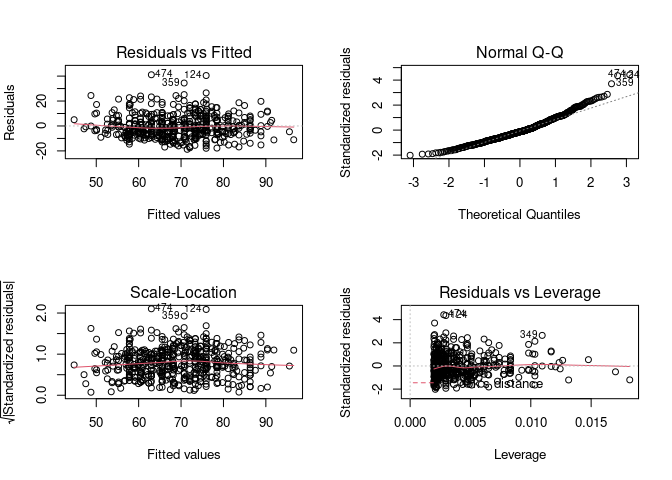
par(mfrow = c(1, 1))
Assumption 2: Normal distribution of the errors
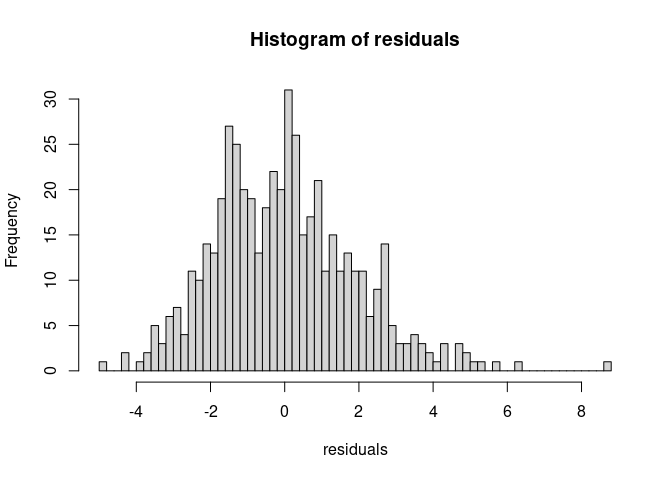
model <- lm(Bicep ~ Shoulder, data = body_data)
residuals <- resid(model)
hist(residuals, 50)
par(mfrow = c(2, 2)) # Split the plotting panel into a 2 x 2 grid
plot(model)
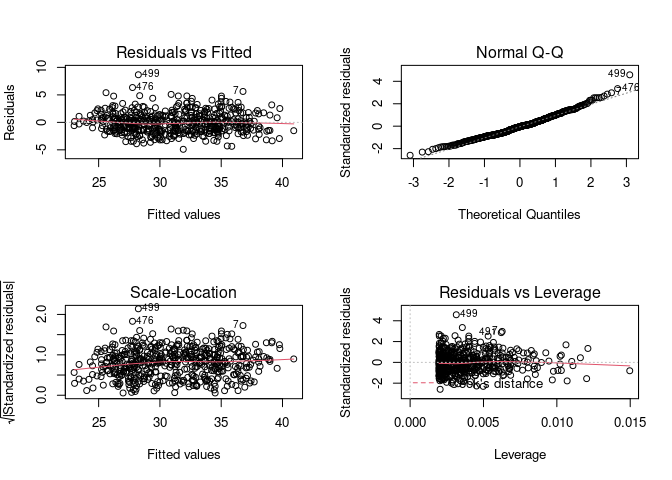
par(mfrow = c(1, 1))
Assumption 3: Homoscedasticity
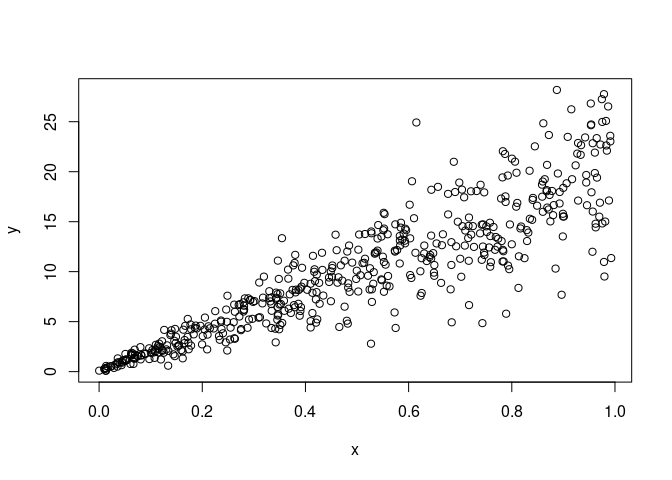
I created a data set with heteroscedasticity. The gambling data below might be a good example as well.
n<-500
errors <-rnorm(n, sd=seq(0.1,5, length.out=n))
x<-runif(n)
x<-sort(x)
y<- (20 * x) + errors
plot(x, y)
fake<-tibble(x=x, y=y)
model <- lm(y~x, data = fake)
The model is still fitted properly!
summary(model)
##
## Call:
## lm(formula = y ~ x, data = fake)
##
## Residuals:
## Min 1Q Median 3Q Max
## -10.2339 -1.3420 0.0339 1.2422 12.6182
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.03805 0.25641 0.148 0.882
## x 19.94614 0.44861 44.463 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 2.891 on 498 degrees of freedom
## Multiple R-squared: 0.7988, Adjusted R-squared: 0.7984
## F-statistic: 1977 on 1 and 498 DF, p-value: < 2.2e-16
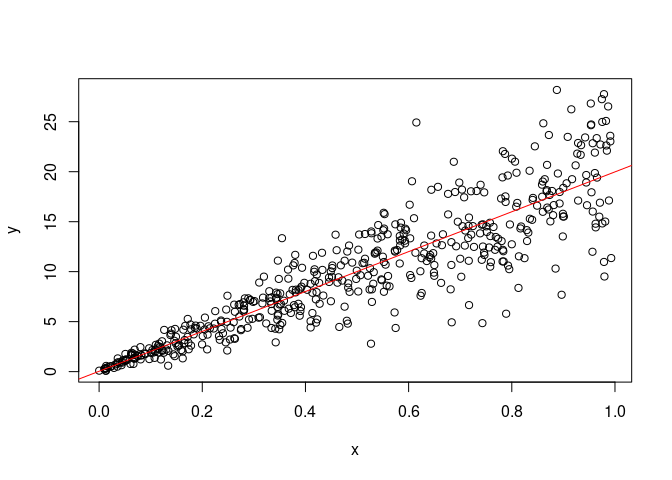
plot(x, y)
abline(model, col='red')
par(mfrow = c(2, 2)) # Split the plotting panel into a 2 x 2 grid
plot(model)
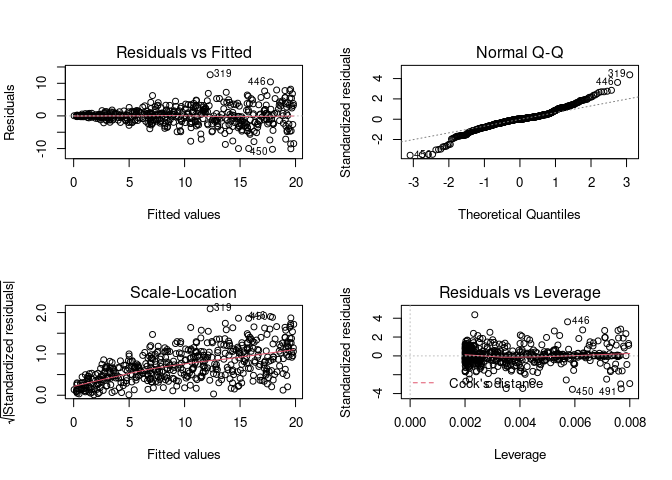
par(mfrow = c(1, 1))
More examples
Heteroscedasticity: Gambling data
The teengamb data frame has 47 rows and 5 columns. A survey was conducted to study teenage gambling in Britain. This data frame contains the following columns:
- sex 0=male, 1=female
- Socioeconomic status score based on parents’ occupation
- Income in pounds per week
- Verbal score in words out of 12 correctly defined
- Gamble expenditure on gambling in pounds per year
library(faraway)
data <- teengamb
head(data)
## sex status income verbal gamble
## 1 1 51 2.00 8 0.0
## 2 1 28 2.50 8 0.0
## 3 1 37 2.00 6 0.0
## 4 1 28 7.00 4 7.3
## 5 1 65 2.00 8 19.6
## 6 1 61 3.47 6 0.1
model<- lm(gamble~income, data=data)
plot(data$income, data$gamble)
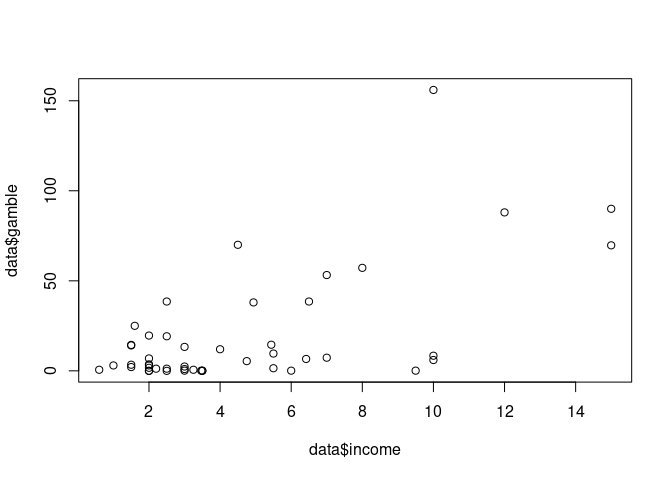
summary(model)
##
## Call:
## lm(formula = gamble ~ income, data = data)
##
## Residuals:
## Min 1Q Median 3Q Max
## -46.020 -11.874 -3.757 11.934 107.120
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -6.325 6.030 -1.049 0.3
## income 5.520 1.036 5.330 3.05e-06 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 24.95 on 45 degrees of freedom
## Multiple R-squared: 0.387, Adjusted R-squared: 0.3734
## F-statistic: 28.41 on 1 and 45 DF, p-value: 3.045e-06
par(mfrow = c(2, 2)) # Split the plotting panel into a 2 x 2 grid
plot(model)
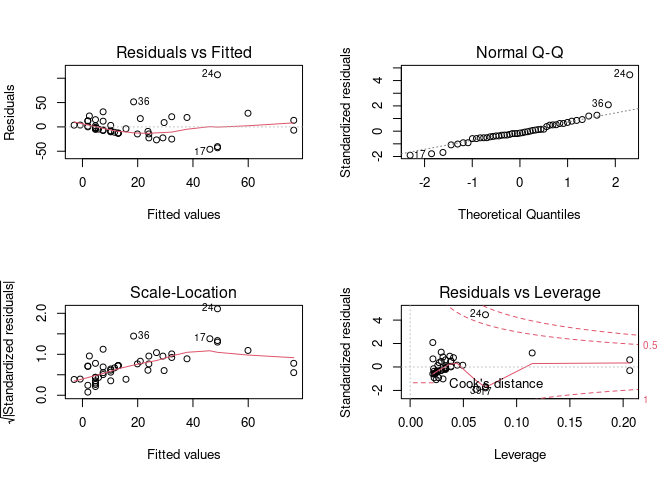
par(mfrow = c(1, 1))
plot(data$status,data$gamble)
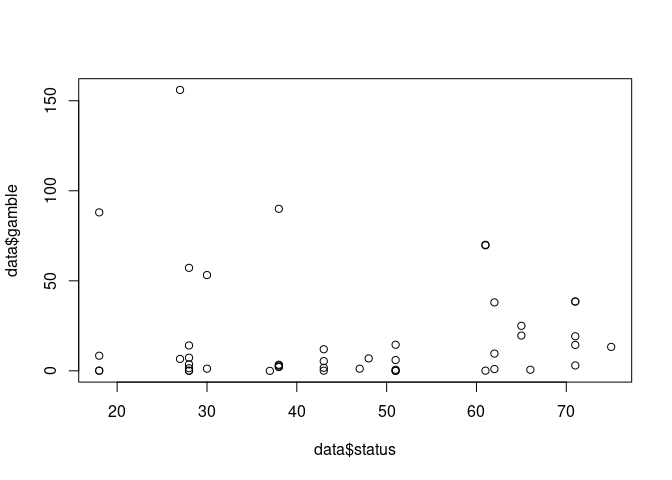
Non-linear relationship: Pima data
The National Institute of Diabetes and Digestive and Kidney Diseases conducted a study on 768 adult female Pima Indians living near Phoenix. The dataset contains the following variables
- Number of times pregnant
- Plasma glucose concentration at 2 hours in an oral glucose tolerance test
- Diastolic blood pressure (mm Hg)
- Triceps skin fold thickness (mm) insulin 2-Hour serum insulin (mu U/ml)
- Body mass index (weight in kg/(height in metres squared))
- Diabetes pedigree function age Age (years)
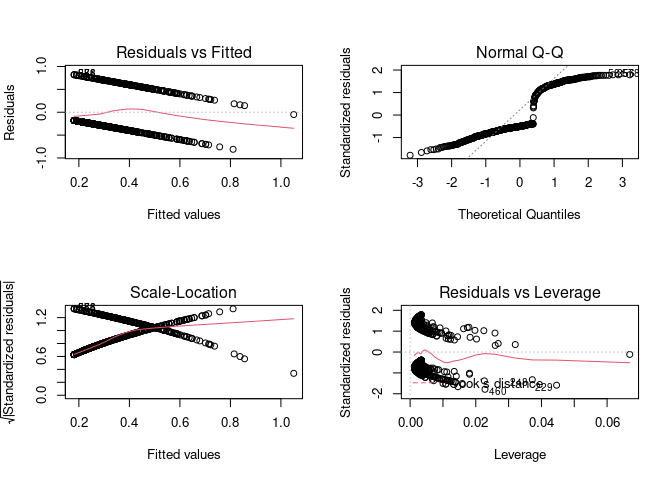
- Test whether the patient shows signs of diabetes (coded 0 if negative, 1 if positive)
data<-pima
model <-lm(test ~ age + insulin, data=data)
par(mfrow = c(2, 2)) # Split the plotting panel into a 2 x 2 grid
plot(model)
par(mfrow = c(1, 1))