Simple Regression
Last Updated: 01, December, 2025 at 19:17
- Before we start…
- Functions we will use
- First example: body data
Before we start…
Download the following data sets:
vik_table_9_2.csvbody.csv
Functions we will use
lm, to fit a linear modelsummary, to get the results from a linear modelanova, to get more results from a linear model (or to compare models)predict, to predict new valuesabline, to plot a regression line
First example: body data
Read Data
library(tidyverse)
## ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
## ✔ dplyr 1.1.4 ✔ readr 2.1.5
## ✔ forcats 1.0.1 ✔ stringr 1.5.2
## ✔ ggplot2 4.0.0 ✔ tibble 3.3.0
## ✔ lubridate 1.9.4 ✔ tidyr 1.3.1
## ✔ purrr 1.1.0
## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
## ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
data <- read_csv('data/body.csv')
## Rows: 507 Columns: 25
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## dbl (25): Biacromial, Biiliac, Bitrochanteric, ChestDepth, ChestDia, ElbowDi...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Run simple regression model
plot(data$Biiliac,data$Height)
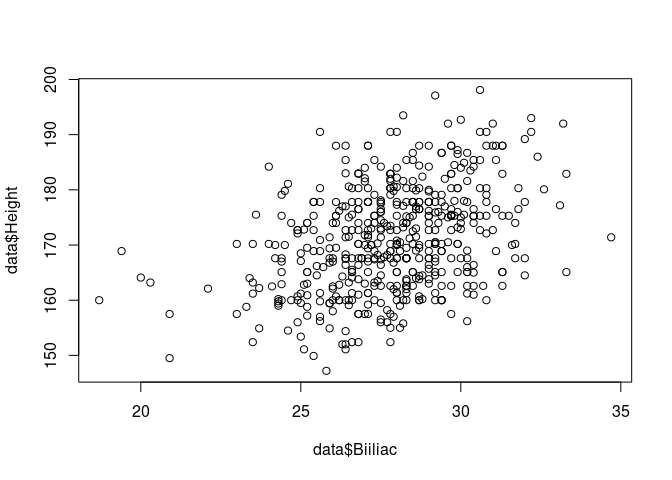 The line
The line result <- lm(Height ~ Biiliac, data = data) fits a linear
model where Height is the dependent variable and Biiliac is the
predictor variable. The line summary(result) provides the results of
the model.
# Note: this syntax makes the predict function work
result <- lm(Height ~ Biiliac, data = data)
# Note: This does not work with the predict function: result <- lm(data$Height ~ data$Biiliac)
summary(result)
##
## Call:
## lm(formula = Height ~ Biiliac, data = data)
##
## Residuals:
## Min 1Q Median 3Q Max
## -20.6424 -6.6158 -0.1255 6.1055 23.7281
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 125.8837 4.8964 25.709 <2e-16 ***
## Biiliac 1.6263 0.1754 9.272 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 8.705 on 505 degrees of freedom
## Multiple R-squared: 0.1455, Adjusted R-squared: 0.1438
## F-statistic: 85.98 on 1 and 505 DF, p-value: < 2.2e-16
If we wish to know how the F-value was calculated we can ask for the anova table.
anova(result)
## Analysis of Variance Table
##
## Response: Height
## Df Sum Sq Mean Sq F value Pr(>F)
## Biiliac 1 6515 6514.6 85.978 < 2.2e-16 ***
## Residuals 505 38264 75.8
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Compare with correlation
Here we use the cor.test function to calculate the correlation between
the two variables. The line correlation$estimate^2 squares the
correlation coefficient to get the R-squared value. This is the same as
the R-squared value from the linear model.
correlation <- cor.test(data$Height, data$Biiliac)
correlation
##
## Pearson's product-moment correlation
##
## data: data$Height and data$Biiliac
## t = 9.2724, df = 505, p-value < 2.2e-16
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## 0.3044541 0.4534453
## sample estimates:
## cor
## 0.3814241
correlation$estimate^2
## cor
## 0.1454843
Get the predicted values
Below, we generate the predicted values for the existing data and for
new data. The line fitted <- fitted.values(result) generates the
predicted values for the existing data. The line
predicted <- predict(result, newdata=new_data) generates the predicted
values for new data.
For the existing independent variable values
fitted <- fitted.values(result)
plot(data$Biiliac,data$Height)
points(data$Biiliac, fitted, col='red')
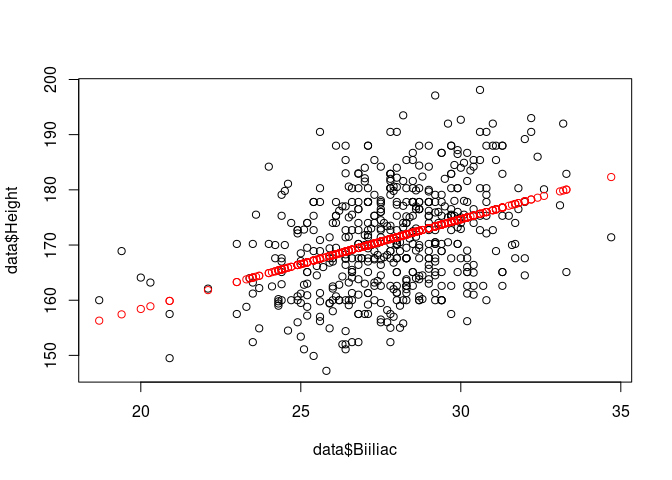
For new independent variable values
More general prediction:
new_independent_values <- c(10,15, 20, 25, 30, 35, 40)
new_data <- data.frame(Biiliac = new_independent_values)
predicted <- predict(result, newdata=new_data)
plot(data$Biiliac, data$Height, col='black')
points(new_independent_values,predicted, col='blue', cex=2, pch=16)
points(data$Biiliac, fitted, col='red')
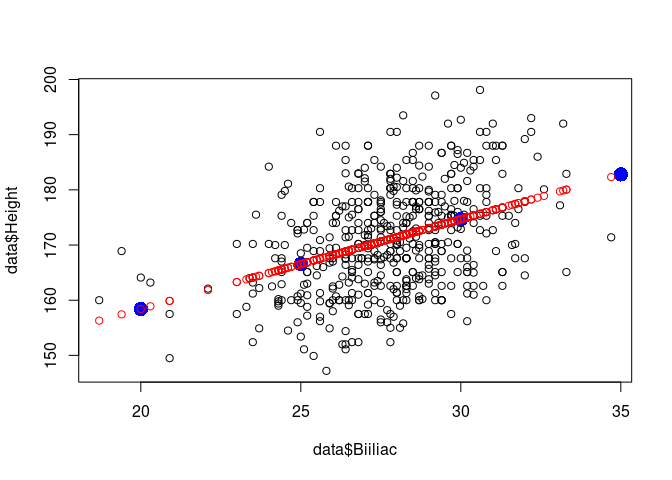
Second example: Vik data
vik_data <- read_csv('data/vik_table_9_2.csv')
## Rows: 12 Columns: 4
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## dbl (4): Person, Y, X1, X2
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
plot(vik_data$X1, vik_data$Y)
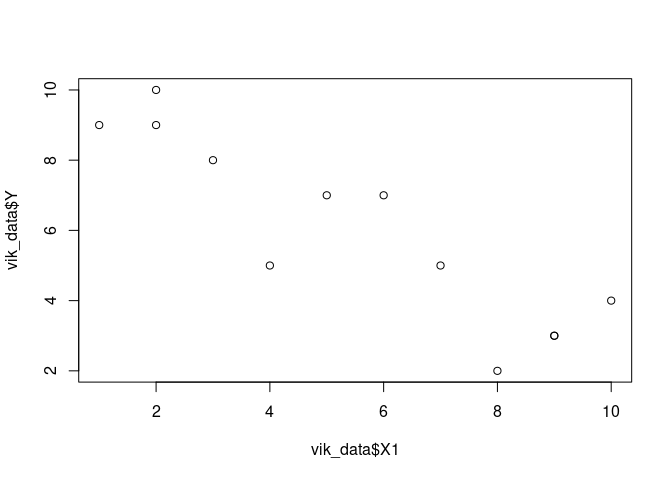
result <- lm(Y ~ X1, data=vik_data)
summary(result)
##
## Call:
## lm(formula = Y ~ X1, data = vik_data)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.1635 -0.3365 0.1121 0.7804 1.4907
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 10.2664 0.7561 13.579 9.07e-08 ***
## X1 -0.7757 0.1208 -6.421 7.63e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.25 on 10 degrees of freedom
## Multiple R-squared: 0.8048, Adjusted R-squared: 0.7853
## F-statistic: 41.23 on 1 and 10 DF, p-value: 7.627e-05
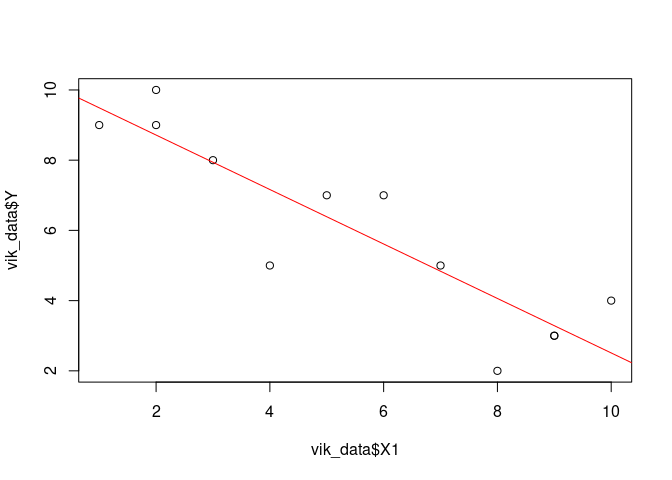
Easier regression line
plot(vik_data$X1, vik_data$Y)
abline(result, col='red')
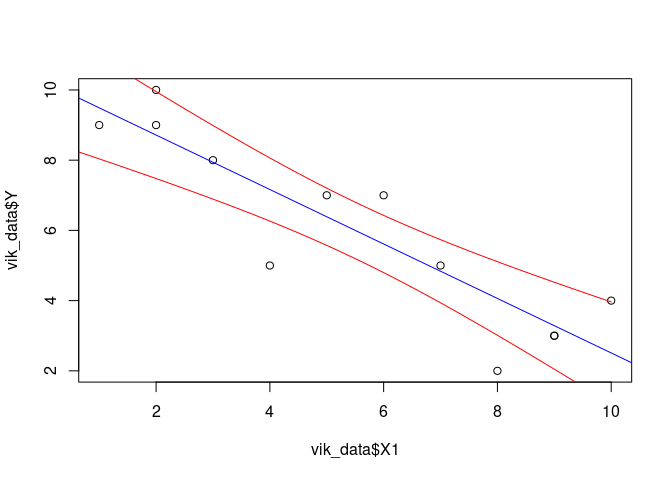
Confidence intervals
Confidence intervals for the predictions
So far, we only generated the best predictions given the model (these are point estimates). However, we can also take into account the uncertainty of the model and generate confidence intervals. Chapter 4 of Faraway, 2009, Linear models with R, provides theory and R code. Here is an example.
The line
predicted <- predict(result, newdata=new_data, interval='confidence')
generates the 95% confidence intervals. The line
lines(new_data$X1, predicted[,2], col='red') plots the lower bound of
the confidence interval, and the line
lines(new_data$X1, predicted[,3], col='red') plots the upper bound of
the confidence interval.
new_data <- data.frame(X1 = seq(0, 10, 0.1))
predicted <- predict(result, newdata=new_data, interval='confidence')
plot(vik_data$X1, vik_data$Y)
lines(new_data$X1, predicted[,2], col='red')
lines(new_data$X1, predicted[,3], col='red')
abline(result, col='blue')
Confidence intervals for the coefficients
We can get the confidence intervals for the estimated coefficients.
confint(result)
## 2.5 % 97.5 %
## (Intercept) 8.581723 11.9509870
## X1 -1.044883 -0.5065185